Research & Projects

-
1
CIMS
My first experiments with a Chemical Ionization Mass Spectrometer
-
2
ECHAMP
Quantification of Peroxy Radicals
-
3
Learning Python
Reflecting on an Application-Based Project
An aspiring atmospheric chemist.
Former member, the
Wood Lab
.
First year PhD student at
CU Boulder Chemistry
in the
Volkamer Lab
.
Learn. Teach. Discover.

My first experiments with a Chemical Ionization Mass Spectrometer
Quantification of Peroxy Radicals
Reflecting on an Application-Based Project
Recently, I've been working on understanding the analytical capabilities of our Chemical Ionization Mass Spectrometer (CIMS) 1 , particularly its sensitivity to various compounds.
The mass spec we use is pretty unique, and it is not the one we learn about in Organic Chemistry $\mathrm{II}$... we're attempting to avoid any fragmentation! (I know, it's wonderful). Essentially, the idea is to bombard everything entering the instrument with the $\ce{I-}$ anion (not electrons like in Electron Ionization ) so that an iodide adduct of the analyte is formed. When we look for a given analyte's signal on the mass spectrum, we eye in on the mass-to-charge ratio of the compound of interest + the mass of $\ce{I-}$. For example, the water peak is found at $m/z\approx145\; (\ce{I-}: 127,\; \ce{H2O}: 18.02)$.
Say you are targeting quantitative analysis of a compound: the signal response of the instrument $S$ is not only dictated by how much of the compound is in your sample but also how sensitive your instrument is to that compound. So, in our lab, we must identify the sensitivity of our instrument for a given compound before we report any concentrations from air samples.
One of the first research topics I worked on recently has been the characterization of nitrogen-containing acids with our CIMS. These acids are important for their presence in some common radical chemistry observed in the atmosphere. Nitrous acid ($\ce{HONO}$) in particular is a source of the hydroxyl radical ($\ce{OH}$), which makes studying it extremely warranted! Our atmosphere is an oxidizing one. This is commonly what atmospheric chemists identify first when describing the Earth's atmosphere. Nothing plays a more important role in all this oxidation than $\ce{OH}$! Nitric acid is an important termination product of the $\ce{HO_x}$ and $\ce{NO_x}$ cycles $(\ce{HO_x}=\ce{HO, HO_2}$ ; $\ce{NO_x}=\ce{NO, HO_2})$ — which include the aforementioned oxidant $\ce{OH}$ as well as dictate $\ce{O3}$ formation and removal. Lastly, peroxynitric acid (PNA, $\ce{HO2NO2}$) acts as a concurrent reservoir of $\ce{HO_x}$ and $\ce{NO_x}$ and in this way, it modifies the oxidative power of the atmosphere. Understanding it too is important.
As mentioned, laboratory-based atmospheric chemistry is concerned with calibration methods. So rather than placing some tubing outside and seeing what signal we get for nitrous, nitric, and peroxynitric acid, we are tasked with producing them ourselves in known quantities. How so?
We treat humidified zero air ($21\% \; \ce{O3} \;\& \; 79\% \; \ce{N2}$) with a $\ce{Hg}$ UV radiation source ($\lambda = 184.9$ nm) to photolyze $\ce{H2O}$ and effectively produce the hydroperoxyl radical ($\ce{HO2}$). This photolysis mechanism goes as follows: $$ \begin{align*} &\text{R1:} \quad &&\ce{H2O +\textit{h}\nu -> OH \text{+} H} \\\ &\text{R2:} &&\ce{H + O2 + \textit{M} -> HO2 \text{+} \it{M}} \end{align*} $$ We then add $\ce{NO_2}$ upstream of the UV source (you can buy a gas cylinder of it). It kicks off a slew of reactions: $$ \begin{align*} &\text{R3a:} \quad &&\ce{OH + NO2 \xrightarrow{\text{preferred}} HO2 + NO} \\\ &\quad \text{R3b:} &&\ce{OH + NO2 + \it{M} -> HNO3 + \it{M}} \\\ &\text{R4:} \quad &&\ce{HO2 + NO2 -> HO2NO2} \\\ &\text{R5:} \quad &&\ce{OH + NO -> HONO} \end{align*} $$ Et voila! All that's needed are the flow rates we used & some rate constants and we can figure out the concentrations of nitrous, nitric, and peroxynitric acid.
Below is a time series of the calibration. The relative humidity (RH; indicated as $\chi_{\ce{H2O}}$) was the independent variable here — allowing us to determine the sensitivities at different RHs. The units of the signal are normalized counts per second. Essentially, it is the count of ions the instrument recognizes normalized to some always-large peaks.
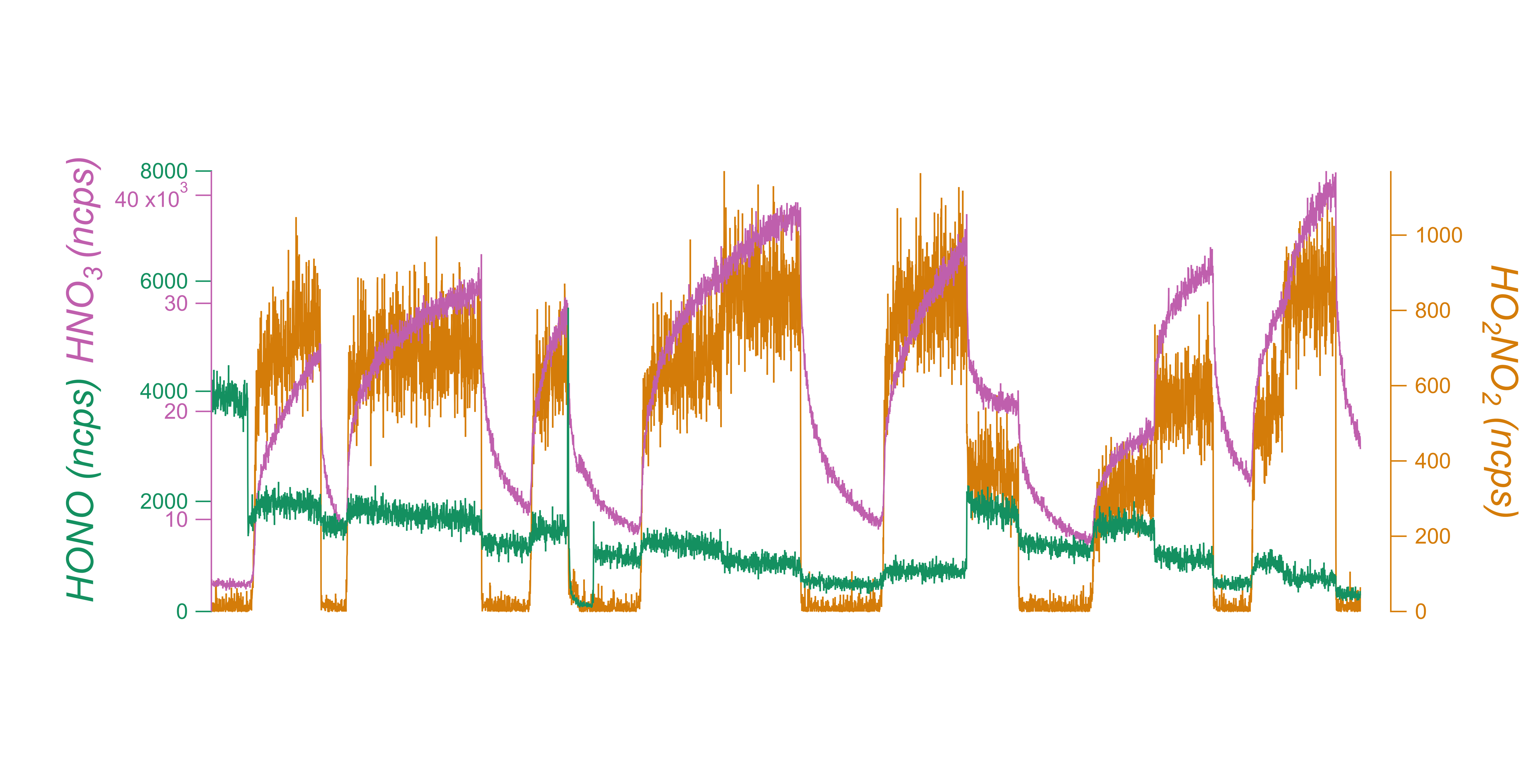
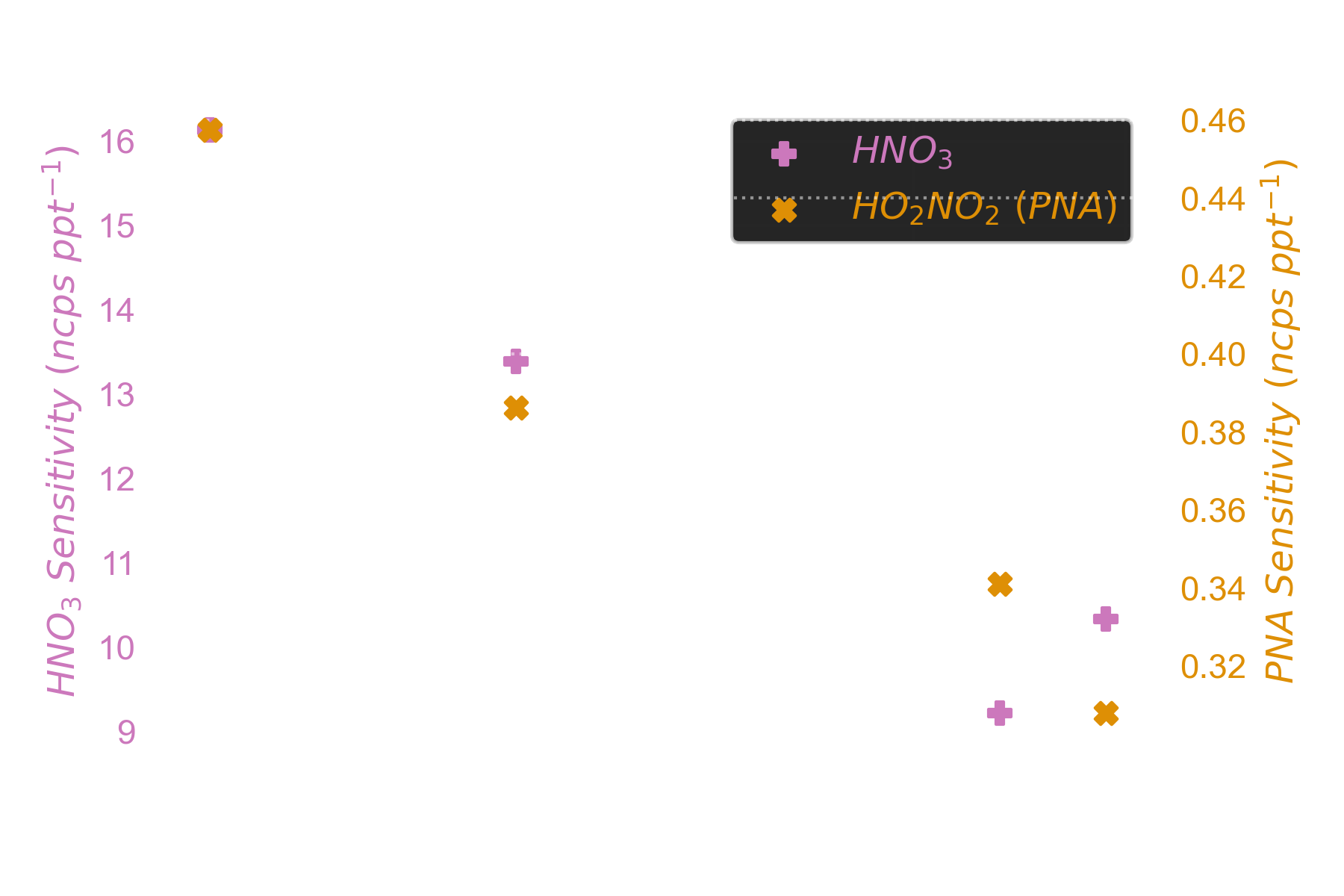
And these are the sensitivities (signal per unit concentration). Here only nitric and peroxynitric acid are shown.
This was one of my first stabs at CIMS work in the lab. Though the steps taken may sound a bit simple (they did to me when we planned out the work) I learned that it wasn't anything of the sort. Our configuration has the CIMS collecting a new mass spectrum every second, $1 \; Hz$. This results in a lot of data (relative to what I'm used to!)— the processing of which requires a lot of meticulous work. Nonetheless, with each peak fit and peak identification, you learn more about the gas phase presence and ion-molecule interactions of the vast assortment of things the mass spectrometer can see. Not only that but the analysis process is eye-opening to the surprising creativity that exists in data analysis. So many chemical behaviors and relationships can be made if one has a command of the data and looks at it with ever-changing perspectives.
Our research group (Ezra Wood, Drexel University) is interested in understanding tropospheric ozone and the chemistry that forms it & removes it. Our main contribution to the understanding of these processes is through quantification of the very reactive peroxy radicals. These reactive species play an important role in ozone production, $P(O_3)$,
Peroxy radicals are difficult to measure using traditional measures. Take carbon dioxide, for contrast. Carbon dioxide is both stable and strongly IR absorbent. All one needs is only a small amount of $CO_2$ in a gas sample for detection. In the case of peroxy radicals, their reactivity makes it difficult to introduce a sample to the instrument for measurement. The solution, then, is to measure that reactivity. We measure the peroxy radicals' reaction with $NO$ to form $NO_2$ (which is something we can measure directly).
Also, these peroxy radicals are in such low concentrations! This 'proxy' reaction would better suit us if there were more molecules of the product we make than the molecules of peroxy radicals to start with. Fortunately, there's a way! By introducing ethane to the reaction system, we don't just double the number of molecules $NO_2$ produced per molecule peroxy radical, we also return another molecule of the peroxy radical! The introduction of ethane starts a chain reaction.
This method is what's called Ethane Chemical Amplification (ECHAMP).
The experimental 'chain length' is the effective number of molecules of $NO_2$ made per molecule of peroxy radical. The larger the chain length (it is variable based on environmental conditions), the more sensitive our instrument is to peroxy radicals. Some of our current work on the ECHAMP surrounds improving this sensitivity — in fact, this was one of my first projects in atmospheric chemistry!
In this paper I have recorded my accounts, lessons, and reflections on learning how to program with Python through an application-based project.
I am a third-year undergraduate student studying chemistry, and I want to focus my work on atmospheric chemistry in my graduate & post-graduate and/or industry research. I love what I learn in this field and every day I see dots of seemingly chaotic and unconnectable complexities get connected. Each new figure showing concentration vs time, oxidation products of terpenes, radiance vs wavenumber, and so on gives more meaning to what’s going on around me. And so I’ve asked myself “How can I do more? How can I be a better chemist? How can I accelerate this learning process so that, sooner rather than later, I too can contribute to further understanding?” This whole paper is a recollection of one of the more important answers I’ve identified to that question: learn Python.
Fortunately, I’ve been exposed to some atmospheric research, specifically that by Dr. Ezra Wood and his group attempting to improve the sensitivity of their peroxy radical sensor. The biggest takeaway from this experience was seeing first-hand how essential computational analysis is. Being able to analyze and organize the hundreds and thousands of data points per day and then evaluate it for all types of trends is the brunt of the research. This process is extensive but critical. No chemist can succeed without strong skills in these areas. And I want to succeed — because, well, I want to contribute to further understanding.
I got to the answer of my aforementioned, driving question by speaking with professors, fellow students, and other researchers... I learned that the most robust and transferrable skill needed to perform these types of data organization, model construction, and computational analyses is to learn how to program — ideally with a language that excels with such tasks. As I said, I chose Python.
Here's how I learned it.
*I didn’t fully learn it. I don’t think I ever will. I just liked how a 5-word sentence sounded to close out the introduction. I feel I’ve ruined that effect now though.
I knew way before I started this process that I wanted to learn Python by completing a project. I figured it would be an exciting way to learn something new as I’d have complementary motivations all at play. I can learn the programming, learn the project, and have a tangible outcome to reflect on. My first problem, then, was to conceive a proper and achievable project idea. I wasn’t sure of the extent to which one project would be more apt to teaching programming basics vs another — let alone the capabilities of programming on its own. Because the ideal subject of said project was something that combined Python and atmospheric chemistry, I asked my (soon to be at the time) atmospheric chemistry professor if he had any ideas/suggestions.
He did! Dr. Wood put me in contact with Dr. Shannon Capps who leads the Atmospheric Modeling group at Drexel (the existence of which was unbeknownst to me at the time). I had never heard of her work or spoke with her before, but she kindly responded to my cold email with enthusiasm and an idea that eventually became the topic of my project. Dr. Capps and I met over zoom not long thereafter, and the plan was set in motion.
The Atmospheric Modeling group is attempting to propagate sensitivities from an outdoor, regional model they have to an indoor model that is written in Python. To do this, they need a specific package to mathematically utilize hyper-dual numbers. This hyper-dual library is currently written in Fortran, so it must be translated to Python. That’s my project.
When Dr. Capps and I first met, one of the first things I told her was that my knowledge of programming and computer science (I didn’t even know what terms to use) was very minimal. I wanted to be clear and up-front that though my ambition would let me take on any project idea she may be able to come up with (this hyper-dual package included), I should probably stick to something manageable and realistic. She appreciated this honesty, because ultimately this project is helping her group too, but did say that it is a realistic goal given the limited background I had. We both agreed that once I could demonstrate a very basic command of Python to Dr. Capps, I’d be given access to the Fortran hyper-dual library and could start working. I was excited to have found a project and ready to start.
One of the perks of this project is that translating code from one language to the next involves relatively little programmatic thinking — the focus is more on common syntax. The programming is already done, I just need to translate it to the proper language. That said, by being exposed to said programming, I can slowly pick it up as I go along. I need to know what exactly I am translating — later this proves to be an important aspect to the project.
After the short turnaround between voicing my interest to Dr. Wood and then the conception of a full project plan, I was enthusiastic about embarking. I learned quickly however that learning to program is full of repeated self-starting and restarting. More than anything, it’s filled with a lot of links. Here’s some words I had journaled at the start:
I never actually used EndNote, but I would say going down that rabbit hole and the many others at that stage were necessary to get started. It was the barrier I needed to overcome, and though a more direct and guided approach would’ve been more efficient, I do believe I learned more this way. Even though I didn’t use a lot of the information I read about, I learned how other people have done similar projects. I invested my time, cleared up my vision a bit, and humbled myself.
I downloaded the Anaconda environment and started with the Jupyter Lab web-based IDE from the Anaconda Navigator. Anaconda, as I learned, is a Python and R programming distribution created for Data Science. The easiest way for beginners to think about it is a big box that contains everything you need to get started. With it comes Python, hundreds of Python libraries (like NumPy, Pandas, SciPy, etc.), and different IDEs to code in. An IDE is an Integrated Development Environment, meaning it is a place where one can write code, execute code, and perform other tasks related to the development of a program. An IDE is different than a regular Code Editor (like TextEdit, Notepad, and Notepad ++) because it allows you to do much more than just write and edit. I quickly found Jupyter Lab to be the best possible IDE to start with because your programs can be executed on the same document as your work. The interface is user-friendly and not overwhelming.
With my understanding of these platforms where it needed to be and the platforms themselves downloaded and installed, I was ready to write and execute my first Python code. Finally!
But how...
Eager and impatient, after finishing a movie on a Friday night, I displayed my MacBook screen on my living room TV and picked a random thumbnail that comes up on YouTube when you search “Python beginner tutorial.” I had Jupyter Lab open on my Mac and “Learn Python in 1 Hour” open on my TV at 12:30 AM Saturday morning... this is how!
I started coding and didn’t stop till I made it to the end. After all of the pauses, replays, and own attempts of following along, it wasn’t until 4:00 AM that I finished. The 1-hour tutorial taught me a lot of the basics very fast, and I was truly surprised at how quickly this language can be picked up. I found it extremely helpful to think of what I was doing in terms of things I already knew. Below is a few of the things I made note of early on that night:
Variables are the same things as the ones in Maple. Or like the ones on the TI-84 calculator. On the calculator when you hated repeatedly typing “$6.67430 \times 10^{-11}$” for the gravitational constant, you’d type it in and then hit the “store” button and store it as “G.” In Python, variables store something in the memory of the computer. Later on, you can use this something in a calculation or an action.
Using information is easy in Python; however, you need to make sure that your “expressions” or, for me (because I think of programming as a maple worksheet) “equations” use things that match up. For example, if you define the variable “happiness_level” to equal input(“Enter your happiness level: “) , then the variable will be equal to something that is within quotes. Anything within quotes is called a string. A string is not a number and therefore you cannot perform the operation: 10 - “happiness_level” . Instead, you must make the type of values in the operations consistent.
Making value types consistent is done with 4 simple functions. First is the int() function which has the ability to seek within a string (something within quotations) and make it an integer number. Next is string() which makes a value a string value (opposite integer). Another useful function is float() which converts values to a floating point number (a number with a decimal). Last is the bool() function which converts a value to a Boolean (binary, true or false).
Getting information is important for learning Python, it’s a very useful way to grow your programming. The two most important commands (I think that’s the word?) for this are input(“ “) and print() . The input() command lets you make a prompt followed by a field that can be used to enter information. This information can then be manipulated by your program. Once manipulated, you can opt to display the results of what the code has done. Displaying these results is achieved by the print() command.
I enjoyed the teaching style of the video creator because he would talk through everything, he typed in a way that I could follow along and code with him. I would learn in the moment and have my own, slightly adapted, code to look back on after the fact.
Here are two of the earliest Python codes of mine:
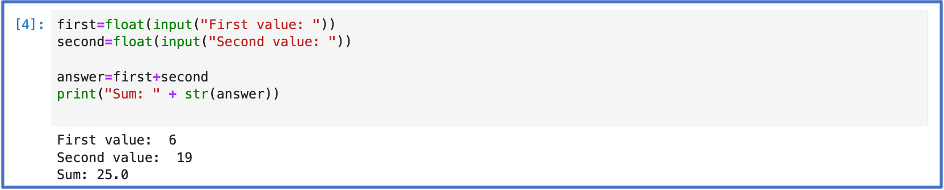

In the first of the two codes above, I told the computer to let me give the program some information. When executed two input boxes appeared. One let me input the first value, and the next asked for the second. The program then gave me the sum. In the second code, I told the computer to print the answer to the logical statement “Python” in “Python for atmospheric chemist.”
There are a whole lot more of such codes in my (no better name than...) “Hello World” Python file.
After graduating from this initial tutorial, I enrolled in a proper online course offered by the same creator of the original video, Mosh Hamdani. I began learning more in-depth about these basic principles and finding new ways to build upon the most basic of concepts.
Here are some more of the early codes from this time:

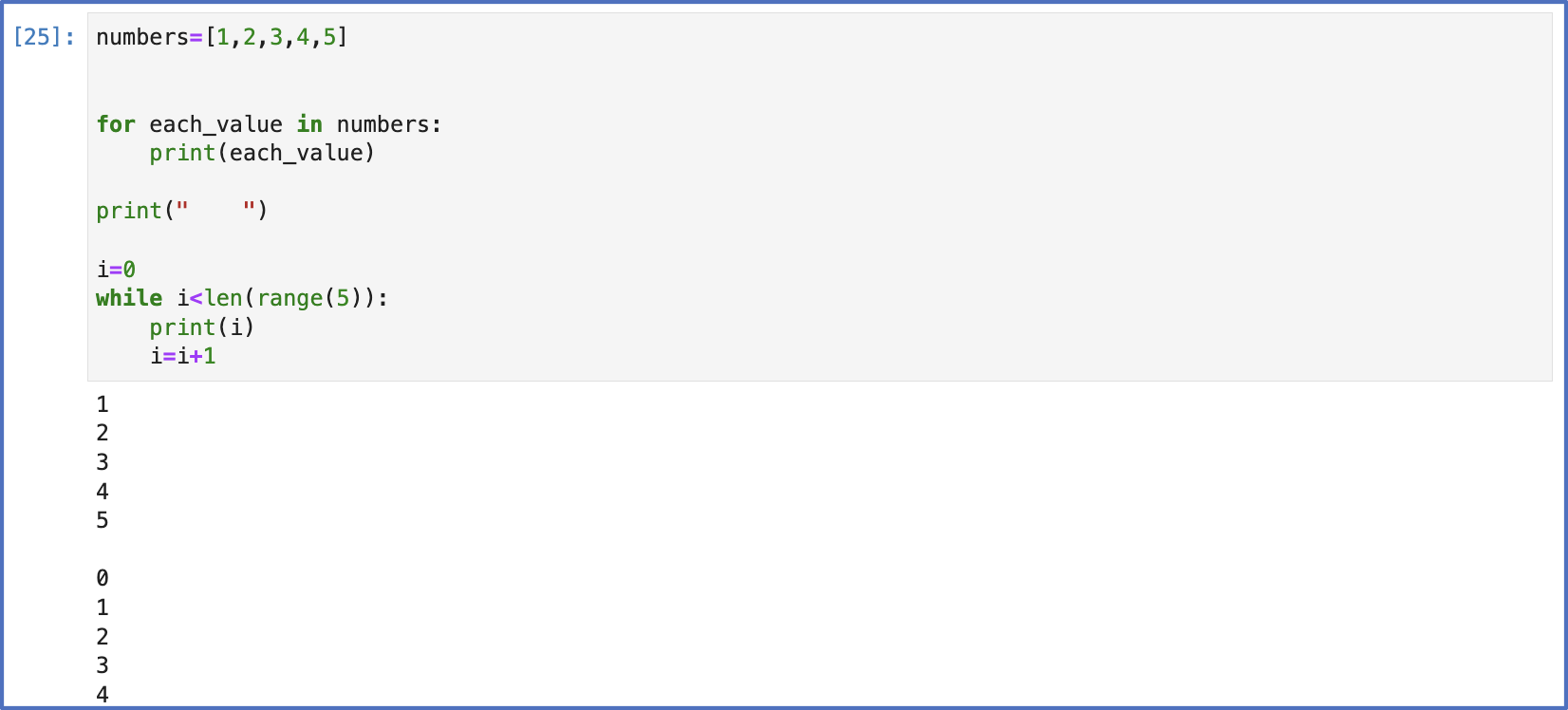
Though (seemingly) simple, the loops discussed above carry with them important principles in Python programming. Having command of loops and understanding iteration is critical to high-level computations in this language. Python is an object-oriented program language, which means its programs heavily rely on the objects themselves. A Python programmer will find that the skill of iterating over objects is a common and essential task; therefore, making and manipulating loops is something I try to practice in different ways.
As you can see, my grasp of the language grew. However, I knew I’d need to start splitting my path away from straight web development (which is what Mosh focuses on) to more computational applications of the language. To do this, I decided I’d try my hand at plotting some data. This was an exciting point for me because I knew I had the rudimentary skills down, and I was able to create my own programs without the direct help of Mosh. That said, a learning curve awaited me. This one however was not like the rest. It was easier this time and I found this encouraging.
I knew that the first step was to accustom myself a bit with what packages are in Python. Aside from knowing about packages from this project itself, I had already read about them here and there when link surfing early in the process. From what I knew, packages are big programs that you can import into your own program to perform tasks that the Python language “default” can’t do (without of course making your program as equally complex as the package). If I wanted to write a relatively small code that would extract data and plot it, I figured I’d need to import a package.
I was right.
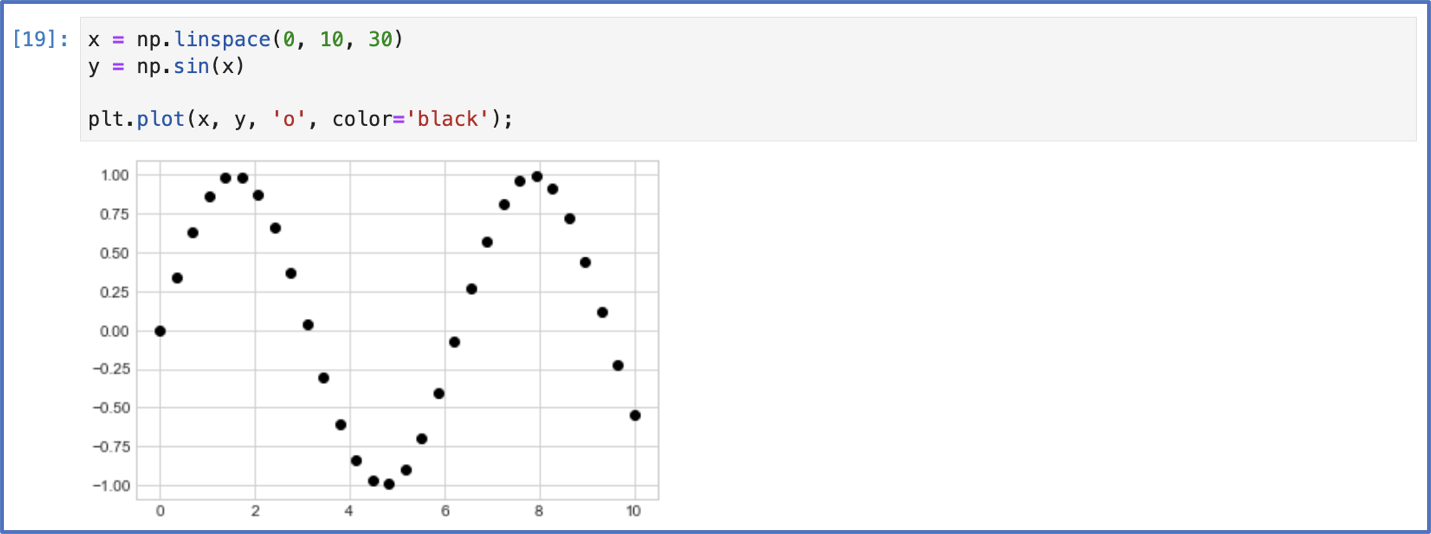
Thanks to a quick google search, I found that the main plotting package on Python is matplotlib. I imported it and another package NumPy like the person on the forum suggested. Then I started step 2... I tried something!
Below is my first plot. $f(x) = sin(x)$:
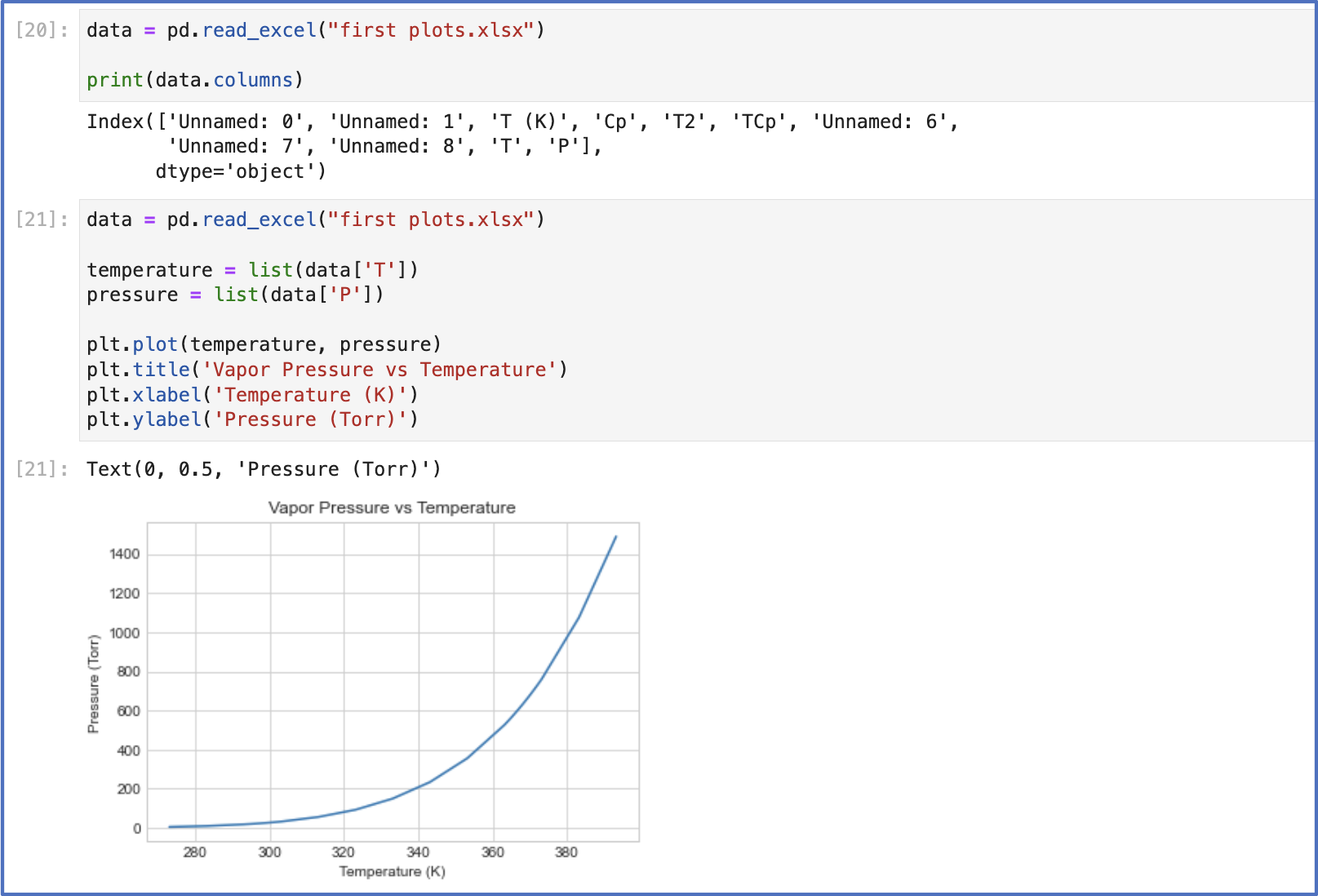
The next task I set for myself was to plot some data from excel. Another serviceable google search showed that to do this, I need to import the Pandas library to extract my data from excel.
Here’s how it went:
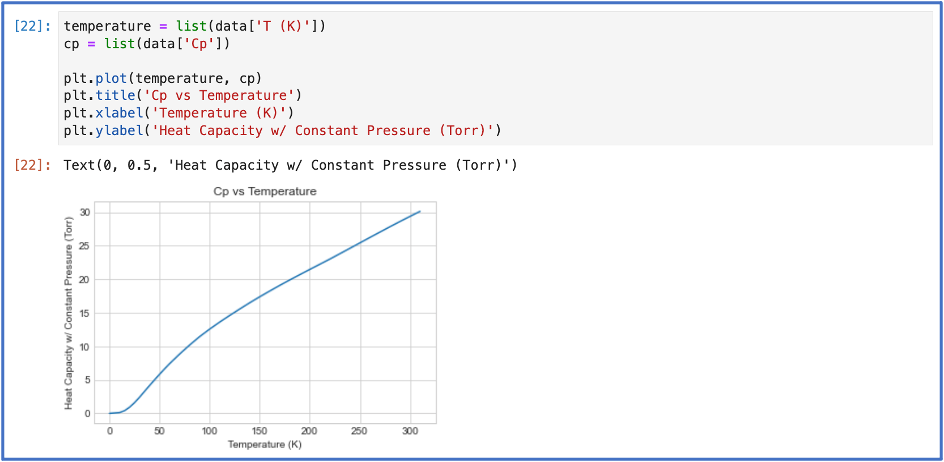
I was having a lot of fun, but I knew that simple plotting wasn’t really challenging me. I wanted to try something a bit more sophisticated, so I decided I would try something that would let me plot, fit data, perform matrix multiplication, and integrate all in one.
I had some data of heat capacity at constant pressure of some sample collected at different temperatures and wanted to find the change in entropy of this sample from 100 K to 300K. The change in entropy can be expressed as: $$ \Delta S = \int \frac{C_p}{T} \; \textit{d}T. $$ I needed to find a functional form of Cp with respect to T. This is where the fitting came to play. Once I found the functional form of Cp, I used another package, SciPy, that could handle my integration. Below is all the code that did this:
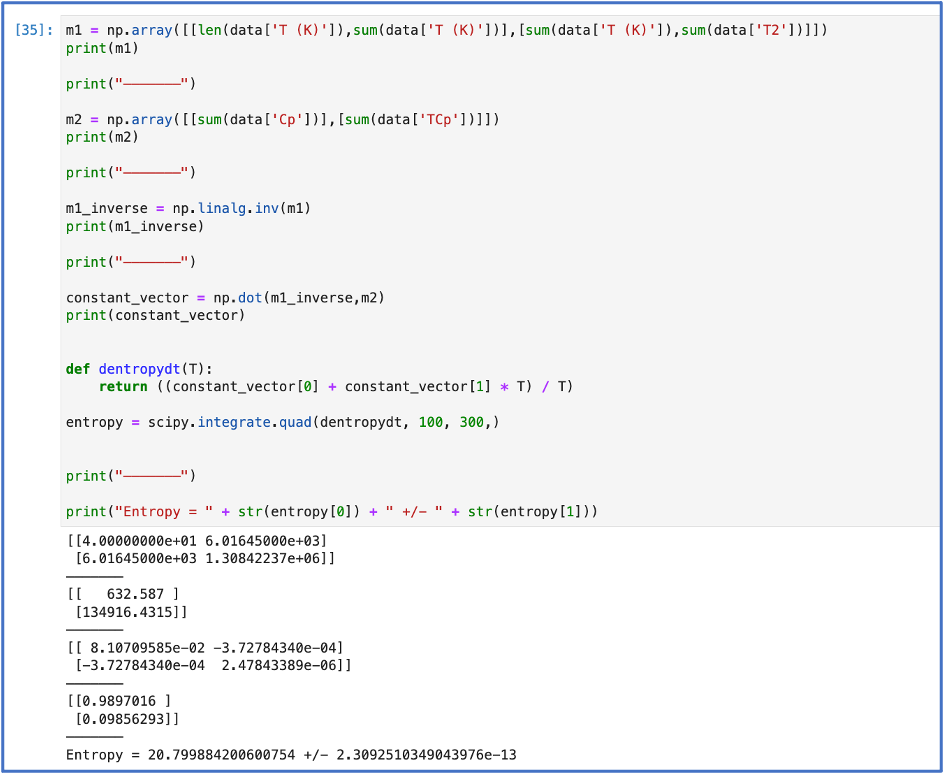
I had found that this was precisely the application of fundamental skills I needed to take the next step. In completing this mini problem, I was learning how to effectively research documentation, utilize the (oh so) important functions in python, manipulate numbers, troubleshoot, and so many other things. I felt ready to begin the hyper-dual library!
At this point in my progress, I had met with Dr. Capps twice since our first meeting and we both felt that I was ready to begin. She helped me get started and guided me in the right direction — starting with classes. To kick off this section, I include a journal entry from my very first hyper-dual library code!
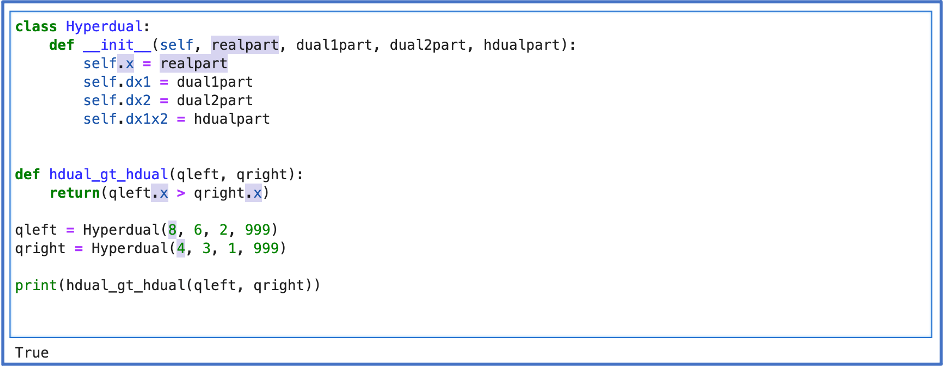
Here, I am very simply replicating a desired use of this library. Obviously, Python doesn’t know how to handle hyper-dual numbers, so with this bit of code, I am allowing python to determine if one hyper-dual number is bigger than another.
I’ve defined some variables “qleft” and “qright” (standing for hyper-dual numbers on the left and right side of the operator) and I am asking the computer to determine if the left-side hyper-dual number is bigger than the right side hyper-dual number.
The first step of this process is to create a class — some “thing” that the computer doesn’t know about that I am teaching the computer to know about. This “thing” is “Hyperdual.” The first step is to tell the computer that a hyper-dual number will be presented to it with 4 ‘parts’: the real part, the first dual part, the second dual part, and the hyper-dual part. I’ve told the computer also how it can isolate each of those parts from any hyper-dual number with which it’s been presented.
After this administrative work, I needed to tell the computer how it can compare hyper-dual numbers based on size. I’ve told it that when given a hyper-dual number, “compare only the first ‘part’ (the real part) of each hyper-dual number — perform this comparison by seeing if the left is greater than the right (>).”
To show the results of the comparison, I’ve printed the function which returns a Boolean (true or false).
In the image shown, all of the highlighted matter is all that corresponds to the real ‘part’ of the hyper-dual numbers. You can see what the function is taking out of the hyper-dual numbers to compare them.
Quickly, I was able to copy & paste and find & replace so that the greater than function became the greater than function, the less than function, the greater than or equal to function, etc. I finished all the logical operators in the library. Now, effectively, one is “able to” compare two hyper-dual numbers!
This was only the tip of (the tip of) the iceberg, however. The list of operations in the Fortran library is extensive and quickly gets complicated.
I moved on next to the arithmetic operators (adding two hyper-dual numbers together for example) and already it got a challenging. One of the main aspects to adding hyper-dual numbers is that you add each respective part of one hyper-dual number to the other hyper-dual number. To make it clear, a hyper-dual number is shown like this:
$$ \begin{align*} &x_{hd} = x_0+x_1 \epsilon_1+x_2 \epsilon_2+x_{12} \epsilon_1 \epsilon_2 \: ;\\\ &\text{where} \: x_1, \: x_2,\: \text{and}\: x_{12} \: \text{are real numbers} \: \text{and} \\ &\epsilon_1 \: \text{and} \: \epsilon_2 \: \text{are dual numbers}. \end{align*} $$
At first, I found it to be an issue that Python functions would only return one object, not independent objects that would each correspond to a part of the hyper-dual number sum. I learned later, however, that keeping the output of my functions inside 1 object is actually better. This is because it gives the computer and the user less to keep track of. Say someone wanted to use the addition function and right after wanted to use that sum and multiply it to another number. In this case, having a singular object as the sum is better. Ultimately, this is true for all cases, but it took me a while to realize this. The next hurdle to overcome was (and partly still is), exactly what object do I want these functions to output?
I went to Dr. Capps for guidance on this whole function output question, and, funny enough, I found out the answer was obvious. It’s important that the output of these arithmetic operations are objects within the “Hyperdual” class I constructed — the point of having a “Hyperdual” class is to formalize a way for the computer to handle these numbers after all. Arriving at this realization, the next step is to consider ways to achieve my functions becoming — succeeding in making this a reality is the big next step and where I am at on this project. The solution can either be class-based where I modify exactly how the “Hyperdual” class operates, or function based where I find a way for my functions to obey the class.
Another problem I’m facing is storing the “Hyperdual” objects into arrays, matrices, and tensors so that one hyper-dual number can be added to a certain set of others simultaneously. A solution to this problem undoubtedly will require iteration... so it’s good that I’ve practiced my loops! Nevertheless, I need to solve the aforementioned problem first and then I can move on to arrays.
My overall take away from the work I have completed so far is very much a positive one. I look back when I originally pitched this idea and I know that there are so many things that I can now do that I couldn’t then. I feel like the skills I have acquired put me in a spot where I am comfortable self-starting other difficult undertaking — specifically those having to do with Python.
My satisfaction however isn’t full... I will continue working on the Python hyper-dual library because I want to complete it and see it through. This “deliverable” is but part of the motivation I originally had to embark on this project, the rest is still standing. The plan was never to complete the hyper-dual library in 10 weeks, so I am not set back by this. But I do feel that I won’t be done until there is no longer more I can do more to build my tool-kit and make contributions to the topic I love.
I owe a gigantic thank you to Dr. Capps for her time and effort. Finding time to work on this project been very difficult despite the immense desire and interest I have for it. Knowing this, I can’t express how lucky I am to have worked with such a committed and generous mentor in Dr. Capps. She made time each weak to meet with me for an hour or more providing one on one guidance. Her direction and confidence in me have truly contributed to the success I have had in growing as a programmer and as a problem solver.
I would also like to thank Dr. Wood for his flexibility and openness for allowing me to work on something so original.
I am proud of the work I have done, and I’m looking ahead excitedly at the rest I still have unfinished.
I am now a full year removed from the completion of this project. Happily, I can say that the skeleton of the Hyper-dual library is finished. It can be accessed here on GitHub ( @atmmod is the GitHub for Dr. Shannon Capp's lab — here's her group's website ).
I owe this whole project so much — this is including the paper I've written documenting the work which has promoted thoughtful reflection. Python has become the most used tool in my toolbox! Seemingly everyday I am writing/contributing a script for some new project, work task, or school assignment.
See the list of Python-related programs I've written!

Cooper, Adam, Shenkiryk, Alexis, Chin, Henry, Morris, Maya, Mehndiratta, Lincoln, Roundtree, Kanuri, Tafuri, Tessa, Slade, Jonathan H.
2024
Zang, Cort L., Willis, Megan D.
2025
Unknown
Hu, Qihua, Moon, Jihye, Kim, Hwajin
2024
Borduas-Dedekind, Nadine, Gemmell, Keighan J., Jayakody, Madushika Madri, Lee, Rickey J. M., Sardena, Claudia, Zala, Sebastian
2024
Samet, Jonathan M., Burke, Thomas A., Goldstein, Bernard D.
2017
Kumie, Abera, Worku, Alemayehu, Tazu, Zelalem, Tefera, Worku, Asfaw, Araya, Boja, Getu, Mekashu, Molla, Siraw, Dawit, Teferra, Solomon, Zacharias, Kristin, Patz, Jonathan, Samet, Jonathan, Berhane, Kiros
2021
Casalnuovo, Dominic A., Cheng, Darren, Flores-Romero, Michel, Montesinos-Castellanos, Alejandro, Jen, Coty N.
2023
Montzka, Stephen A., Dutton, Geoff S., Yu, Pengfei, Ray, Eric, Portmann, Robert W., Daniel, John S., Kuijpers, Lambert, Hall, Brad D., Mondeel, Debra, Siso, Carolina, Nance, J. David, Rigby, Matt, Manning, Alistair J., Hu, Lei, Moore, Fred, Miller, Ben R., Elkins, James W.
2018
Lindsay, Andrew J., Weesner, Brigitte M., Banecker, Kyle, Feinman, Lee V., Long, Russell W., Landis, Matthew S., Wood, Ezra C.
2024
Ranney, April P., and Ziemann, Paul J.
2016
Chance, Rosie, Tinel, Liselotte, Sarkar, Amit, Sinha, Alok K., Mahajan, Anoop S., Chacko, Racheal, Sabu, P., Roy, Rajdeep, Jickells, Tim D., Stevens, David P., Wadley, Martin, Carpenter, Lucy J.
2020
Zappal, C. J., Asher, W. E., Jessup, A. T., Klinke, J., Long, S. R.
2013
Chang, Wonil, Heikes, Brian G., Lee, Meehye
2004
Gálvez, Óscar, Baeza-Romero, M. Teresa, Sanz, Mikel, Pacios, Luis F.
2016
Brown, Lucy V., Pound, Ryan J., Ives, Lyndsay S., Jones, Matthew R., Andrews, Stephen J., Carpenter, Lucy J.
2024
M. Prophet, Alexander, Polley, Kritanjan, Berkel, Gary J. Van, T. Limmer, David, R. Wilson, Kevin
2024
Seinfeld, John H., Pandis, Spyros N.
2016
Unknown
2018
Pound, Ryan J., Sherwen, Tomás, Helmig, Detlev, Carpenter, Lucy J., Evans, Mat J.
2020
Conley, Stephen A., Faloona, Ian C., Lenschow, Donald H., Campos, Teresa, Heizer, Clifford, Weinheimer, Andrew, Cantrell, Christopher A., Mauldin, Roy L., Hornbrook, Rebecca S., Pollack, Ilana, Bandy, Alan
2011
Chiu, Randall, Obersteiner, Florian, Franchin, Alessandro, Campos, Teresa, Bailey, Adriana, Webster, Christopher, Zahn, Andreas, Volkamer, Rainer
2024
Iyer, Siddharth, Rissanen, Matti P., Valiev, Rashid, Barua, Shawon, Krechmer, Jordan E., Thornton, Joel, Ehn, Mikael, Kurtén, Theo
2021
DeVault, Marla P., Ziola, Anna C., Ziemann, Paul J.
2022
Chance, Rosie, Tinel, Liselotte, Sarkar, Amit, Sinha, Alok K., Mahajan, Anoop S., Chacko, Racheal, Sabu, P., Roy, Rajdeep, Jickells, Tim D., Stevens, David P., Wadley, Martin, Carpenter, Lucy J.
2020
Zappal, C. J., Asher, W. E., Jessup, A. T., Klinke, J., Long, S. R.
2013
Chang, Wonil, Heikes, Brian G., Lee, Meehye
2004
Gálvez, Óscar, Baeza-Romero, M. Teresa, Sanz, Mikel, Pacios, Luis F.
2016
Brown, Lucy V., Pound, Ryan J., Ives, Lyndsay S., Jones, Matthew R., Andrews, Stephen J., Carpenter, Lucy J.
2024
M. Prophet, Alexander, Polley, Kritanjan, Berkel, Gary J. Van, T. Limmer, David, R. Wilson, Kevin
2024
Seinfeld, John H., Pandis, Spyros N.
2016
Unknown
2018
Helmig, D., Lang, E. K., Bariteau, L., Boylan, P., Fairall, C. W., Ganzeveld, L., Hare, J. E., Hueber, J., Pallandt, M.
2012
Wolfe, Glenn M., Kawa, S. Randy, Hanisco, Thomas F., Hannun, Reem A., Newman, Paul A., Swanson, Andrew, Bailey, Steve, Barrick, John, Thornhill, K. Lee, Diskin, Glenn, DiGangi, Josh, Nowak, John B., Sorenson, Carl, Bland, Geoffrey, Yungel, James K., Swenson, Craig A.
2018
Bariteau, L, Helmig, D, Fairall, C W, Hare, J E, Hueber, J, Lang, E K
2010
Sarwar, Golam, Kang, Daiwen, Foley, Kristen, Schwede, Donna, Gantt, Brett, Mathur, Rohit
2016
Pound, Ryan J., Sherwen, Tomás, Helmig, Detlev, Carpenter, Lucy J., Evans, Mat J.
2020
Torrence, Christopher, Compo, Gilbert P.
1998
Ridley, B. A., Grahek, F. E., Walega, J. G.
1992
Sanchez, Javier, Tanner, David J., Chen, Dexian, Huey, L. Gregory, Ng, Nga L.
2016
Iyer, Siddharth, Lopez-Hilfiker, Felipe, Lee, Ben H., Thornton, Joel A., Kurtén, Theo
2016
Wang, Mingyi, He, Xu-Cheng, Finkenzeller, Henning, Iyer, Siddharth, Chen, Dexian, Shen, Jiali, Simon, Mario, Hofbauer, Victoria, Kirkby, Jasper, Curtius, Joachim, Maier, Norbert, Kurtén, Theo, Worsnop, Douglas R., Kulmala, Markku, Rissanen, Matti, Volkamer, Rainer, Tham, Yee Jun, Donahue, Neil M., Sipilä, Mikko
2021
He, Xu-Cheng, Shen, Jiali, Iyer, Siddharth, Juuti, Paxton, Zhang, Jiangyi, Koirala, Mrisha, Kytökari, Mikko M., Worsnop, Douglas R., Rissanen, Matti, Kulmala, Markku, Maier, Norbert M., Mikkilä, Jyri, Sipilä, Mikko, Kangasluoma, Juha
2023
Song, Mengdi, He, Shuyu, Li, Xin, Liu, Ying, Lou, Shengrong, Lu, Sihua, Zeng, Limin, Zhang, Yuanhang
2024
Bertram, T. H., Kimmel, J. R., Crisp, T. A., Ryder, O. S., Yatavelli, R. L. N., Thornton, J. A., Cubison, M. J., Gonin, M., Worsnop, D. R.
2011
Lee, Ben H., Lopez-Hilfiker, Felipe D., Mohr, Claudia, Kurtén, Theo, Worsnop, Douglas R., Thornton, Joel A.
2014
Platt, Ulrich, Stutz, Jochen
2008
Volkamer, Rainer, Molina, Luisa T., Molina, Mario J., Shirley, Terry, Brune, William H.
2005
Chan Miller, Christopher, Jacob, Daniel J., Marais, Eloise A., Yu, Karen, Travis, Katherine R., Kim, Patrick S., Fisher, Jenny A., Zhu, Lei, Wolfe, Glenn M., Hanisco, Thomas F., Keutsch, Frank N., Kaiser, Jennifer, Min, Kyung-Eun, Brown, Steven S., Washenfelder, Rebecca A., González Abad, Gonzalo, Chance, Kelly
2017
Tan, Yi, Perri, Mark J., Seitzinger, Sybil P., Turpin, Barbara J.
2009
Heald, Colette L., Jacob, Daniel J., Park, Rokjin J., Russell, Lynn M., Huebert, Barry J., Seinfeld, John H., Liao, Hong, Weber, Rodney J.
2005
Mahajan, Anoop S., Prados-Roman, Cristina, Hay, Timothy D., Lampel, Johannes, Pöhler, Denis, Groβmann, Katja, Tschritter, Jens, Frieß, Udo, Platt, Ulrich, Johnston, Paul, Kreher, Karin, Wittrock, Folkard, Burrows, John P., Plane, John M.C., Saiz-Lopez, Alfonso
2014
Lerot, C., Stavrakou, T., De Smedt, I., Müller, J.-F., Van Roozendael, M.
2010
Coburn, S., Ortega, I., Thalman, R., Blomquist, B., Fairall, C. W., Volkamer, R.
2014
Myriokefalitakis, S, Vrekoussis, M, Tsigaridis, K, Wittrock, F, Richter, A, Burrows, J P, Kanakidou, M
2008
Lerot, Christophe, Hendrick, François, Van Roozendael, Michel, Alvarado, Leonardo M. A., Richter, Andreas, De Smedt, Isabelle, Theys, Nicolas, Vlietinck, Jonas, Yu, Huan, Van Gent, Jeroen, Stavrakou, Trissevgeni, Müller, Jean-François, Valks, Pieter, Loyola, Diego, Irie, Hitoshi, Kumar, Vinod, Wagner, Thomas, Schreier, Stefan F., Sinha, Vinayak, Wang, Ting, Wang, Pucai, Retscher, Christian
2021
Volkamer, R., Baidar, S., Campos, T. L., Coburn, S., DiGangi, J. P., Dix, B., Eloranta, E. W., Koenig, T. K., Morley, B., Ortega, I., Pierce, B. R., Reeves, M., Sinreich, R., Wang, S., Zondlo, M. A., Romashkin, P. A.
2015
Chiu, R., Tinel, L., Gonzalez, L., Ciuraru, R., Bernard, F., George, C., Volkamer, R.
2017
Wittrock, Folkard, Richter, Andreas, Oetjen, Hilke, Burrows, John P., Kanakidou, Maria, Myriokefalitakis, Stelios, Volkamer, Rainer, Beirle, Steffen, Platt, Ulrich, Wagner, Thomas
2006
Volkamer, Rainer, Molina, Luisa T., Molina, Mario J., Shirley, Terry, Brune, William H.
2005
Chan Miller, Christopher, Jacob, Daniel J., Marais, Eloise A., Yu, Karen, Travis, Katherine R., Kim, Patrick S., Fisher, Jenny A., Zhu, Lei, Wolfe, Glenn M., Hanisco, Thomas F., Keutsch, Frank N., Kaiser, Jennifer, Min, Kyung-Eun, Brown, Steven S., Washenfelder, Rebecca A., González Abad, Gonzalo, Chance, Kelly
2017
Mahajan, Anoop S., Prados-Roman, Cristina, Hay, Timothy D., Lampel, Johannes, Pöhler, Denis, Groβmann, Katja, Tschritter, Jens, Frieß, Udo, Platt, Ulrich, Johnston, Paul, Kreher, Karin, Wittrock, Folkard, Burrows, John P., Plane, John M.C., Saiz-Lopez, Alfonso
2014
Lerot, C., Stavrakou, T., De Smedt, I., Müller, J.-F., Van Roozendael, M.
2010
Coburn, S., Ortega, I., Thalman, R., Blomquist, B., Fairall, C. W., Volkamer, R.
2014
Thalman, R., Baeza-Romero, M. T., Ball, S. M., Borrás, E., Daniels, M. J. S., Goodall, I. C. A., Henry, S. B., Karl, T., Keutsch, F. N., Kim, S., Mak, J., Monks, P. S., Muñoz, A., Orlando, J., Peppe, S., Rickard, A. R., Ródenas, M., Sánchez, P., Seco, R., Su, L., Tyndall, G., Vázquez, M., Vera, T., Waxman, E., Volkamer, R.
2015
Myriokefalitakis, S, Vrekoussis, M, Tsigaridis, K, Wittrock, F, Richter, A, Burrows, J P, Kanakidou, M
2008
Volkamer, R., Baidar, S., Campos, T. L., Coburn, S., DiGangi, J. P., Dix, B., Eloranta, E. W., Koenig, T. K., Morley, B., Ortega, I., Pierce, B. R., Reeves, M., Sinreich, R., Wang, S., Zondlo, M. A., Romashkin, P. A.
2015
Wittrock, Folkard, Richter, Andreas, Oetjen, Hilke, Burrows, John P., Kanakidou, Maria, Myriokefalitakis, Stelios, Volkamer, Rainer, Beirle, Steffen, Platt, Ulrich, Wagner, Thomas
2006
Ortega, I., Koenig, T., Sinreich, R., Thomson, D., Volkamer, R.
2015
Kaiser, J., Wolfe, G. M., Min, K. E., Brown, S. S., Miller, C. C., Jacob, D. J., deGouw, J. A., Graus, M., Hanisco, T. F., Holloway, J., Peischl, J., Pollack, I. B., Ryerson, T. B., Warneke, C., Washenfelder, R. A., Keutsch, F. N.
2015
Chan Miller, Christopher, Jacob, Daniel J., González Abad, Gonzalo, Chance, Kelly
2016
Vrekoussis, M., Wittrock, F., Richter, A., Burrows, J. P.
2010
Vrekoussis, M, Wittrock, F, Richter, A, Burrows, J P, Cho, Cho
2009
Waxman, Eleanor M., Dzepina, Katja, Ervens, Barbara, Lee‐Taylor, Julia, Aumont, Bernard, Jimenez, Jose L., Madronich, Sasha, Volkamer, Rainer
2013
Thalman, R., Baeza-Romero, M. T., Ball, S. M., Borrás, E., Daniels, M. J. S., Goodall, I. C. A., Henry, S. B., Karl, T., Keutsch, F. N., Kim, S., Mak, J., Monks, P. S., Muñoz, A., Orlando, J., Peppe, S., Rickard, A. R., Ródenas, M., Sánchez, P., Seco, R., Su, L., Tyndall, G., Vázquez, M., Vera, T., Waxman, E., Volkamer, R.
2015
Volkamer, R., Baidar, S., Campos, T. L., Coburn, S., DiGangi, J. P., Dix, B., Eloranta, E. W., Koenig, T. K., Morley, B., Ortega, I., Pierce, B. R., Reeves, M., Sinreich, R., Wang, S., Zondlo, M. A., Romashkin, P. A.
2015
Chiu, R., Tinel, L., Gonzalez, L., Ciuraru, R., Bernard, F., George, C., Volkamer, R.
2017
Song, Mengdi, He, Shuyu, Li, Xin, Liu, Ying, Lou, Shengrong, Lu, Sihua, Zeng, Limin, Zhang, Yuanhang
2024
Thalman, R., Volkamer, R.
2010
Ervens, B., Volkamer, R.
2010
Kim, Dongwook, Cho, Changmin, Jeong, Seokhan, Lee, Soojin, Nault, Benjamin A., Campuzano-Jost, Pedro, Day, Douglas A., Schroder, Jason C., Jimenez, Jose L., Volkamer, Rainer, Blake, Donald R., Wisthaler, Armin, Fried, Alan, DiGangi, Joshua P., Diskin, Glenn S., Pusede, Sally E., Hall, Samuel R., Ullmann, Kirk, Huey, L. Gregory, Tanner, David J., Dibb, Jack, Knote, Christoph J., Min, Kyung-Eun
2022
Peng, Zhe, Jimenez, Jose L.
2019
Forster, Piers M., Smith, Christopher J., Walsh, Tristram, Lamb, William F., Lamboll, Robin, Hauser, Mathias, Ribes, Aurélien, Rosen, Debbie, Gillett, Nathan, Palmer, Matthew D., Rogelj, Joeri, von Schuckmann, Karina, Seneviratne, Sonia I., Trewin, Blair, Zhang, Xuebin, Allen, Myles, Andrew, Robbie, Birt, Arlene, Borger, Alex, Boyer, Tim, Broersma, Jiddu A., Cheng, Lijing, Dentener, Frank, Friedlingstein, Pierre, Gutiérrez, José M., Gütschow, Johannes, Hall, Bradley, Ishii, Masayoshi, Jenkins, Stuart, Lan, Xin, Lee, June-Yi, Morice, Colin, Kadow, Christopher, Kennedy, John, Killick, Rachel, Minx, Jan C., Naik, Vaishali, Peters, Glen P., Pirani, Anna, Pongratz, Julia, Schleussner, Carl-Friedrich, Szopa, Sophie, Thorne, Peter, Rohde, Robert, Rojas Corradi, Maisa, Schumacher, Dominik, Vose, Russell, Zickfeld, Kirsten, Masson-Delmotte, Valérie, Zhai, Panmao
2023
Callahan, Christopher W., Mankin, Justin S.
2022
Zhang, Kai, Zhang, Wentao, Wan, Hui, Rasch, Philip J., Ghan, Steven J., Easter, Richard C., Shi, Xiangjun, Wang, Yong, Wang, Hailong, Ma, Po-Lun, Zhang, Shixuan, Sun, Jian, Burrows, Susannah M., Shrivastava, Manish, Singh, Balwinder, Qian, Yun, Liu, Xiaohong, Golaz, Jean-Christophe, Tang, Qi, Zheng, Xue, Xie, Shaocheng, Lin, Wuyin, Feng, Yan, Wang, Minghuai, Yoon, Jin-Ho, Leung, L. Ruby
2022
Coburn, S., Ortega, I., Thalman, R., Blomquist, B., Fairall, C. W., Volkamer, R.
2014
Thalman, R., Baeza-Romero, M. T., Ball, S. M., Borrás, E., Daniels, M. J. S., Goodall, I. C. A., Henry, S. B., Karl, T., Keutsch, F. N., Kim, S., Mak, J., Monks, P. S., Muñoz, A., Orlando, J., Peppe, S., Rickard, A. R., Ródenas, M., Sánchez, P., Seco, R., Su, L., Tyndall, G., Vázquez, M., Vera, T., Waxman, E., Volkamer, R.
2015
Lerot, Christophe, Hendrick, François, Van Roozendael, Michel, Alvarado, Leonardo M. A., Richter, Andreas, De Smedt, Isabelle, Theys, Nicolas, Vlietinck, Jonas, Yu, Huan, Van Gent, Jeroen, Stavrakou, Trissevgeni, Müller, Jean-François, Valks, Pieter, Loyola, Diego, Irie, Hitoshi, Kumar, Vinod, Wagner, Thomas, Schreier, Stefan F., Sinha, Vinayak, Wang, Ting, Wang, Pucai, Retscher, Christian
2021
Volkamer, R., Baidar, S., Campos, T. L., Coburn, S., DiGangi, J. P., Dix, B., Eloranta, E. W., Koenig, T. K., Morley, B., Ortega, I., Pierce, B. R., Reeves, M., Sinreich, R., Wang, S., Zondlo, M. A., Romashkin, P. A.
2015
Chiu, R., Tinel, L., Gonzalez, L., Ciuraru, R., Bernard, F., George, C., Volkamer, R.
2017
Song, Mengdi, He, Shuyu, Li, Xin, Liu, Ying, Lou, Shengrong, Lu, Sihua, Zeng, Limin, Zhang, Yuanhang
2024
Thalman, R., Volkamer, R.
2010
Ervens, B., Volkamer, R.
2010
Kim, Dongwook, Cho, Changmin, Jeong, Seokhan, Lee, Soojin, Nault, Benjamin A., Campuzano-Jost, Pedro, Day, Douglas A., Schroder, Jason C., Jimenez, Jose L., Volkamer, Rainer, Blake, Donald R., Wisthaler, Armin, Fried, Alan, DiGangi, Joshua P., Diskin, Glenn S., Pusede, Sally E., Hall, Samuel R., Ullmann, Kirk, Huey, L. Gregory, Tanner, David J., Dibb, Jack, Knote, Christoph J., Min, Kyung-Eun
2022
Myriokefalitakis, S., Vrekoussis, M., Tsigaridis, K., Wittrock, F., Richter, A., Brühl, C., Volkamer, R., Burrows, J. P., Kanakidou, M.
2008
Peng, Zhe, Jimenez, Jose L.
2019
Woodward-Massey, Robert, Sommariva, Roberto, Whalley, Lisa K., Cryer, Danny R., Ingham, Trevor, Bloss, William J., Ball, Stephen M., Cox, Sam, Lee, James D., Reed, Chris P., Crilley, Leigh R., Kramer, Louisa J., Bandy, Brian J., Forster, Grant L., Reeves, Claire E., Monks, Paul S., Heard, Dwayne E.
2023
Baier, Bianca C., Brune, William H., Miller, David O., Blake, Donald, Long, Russell, Wisthaler, Armin, Cantrell, Christopher, Fried, Alan, Heikes, Brian, Brown, Steven, McDuffie, Erin, Flocke, Frank, Apel, Eric, Kaser, Lisa, Weinheimer, Andrew
2017
Lindsay, Andrew J., Anderson, Daniel C., Wernis, Rebecca A., Liang, Yutong, Goldstein, Allen H., Herndon, Scott C., Roscioli, Joseph R., Dyroff, Christoph, Fortner, Ed C., Croteau, Philip L., Majluf, Francesca, Krechmer, Jordan E., Yacovitch, Tara I., Knighton, Walter B., Wood, Ezra C.
2022
US EPA, Region 1
Woodward-Massey, Robert, Sommariva, Roberto, Whalley, Lisa K., Cryer, Danny R., Ingham, Trevor, Bloss, William J., Ball, Stephen M., Lee, James D., Reed, Chris P., Crilley, Leigh R., Kramer, Louisa J., Bandy, Brian J., Forster, Grant L., Reeves, Claire E., Monks, Paul S., Heard, Dwayne E.
2022
Iyer, Siddharth, Reiman, Heidi, Møller, Kristian H., Rissanen, Matti P., Kjaergaard, Henrik G., Kurtén, Theo
2018
Muralikrishna, Iyyanki V., Manickam, Valli
2017
US EPA, OAR
2016
Michalski, G., Bhattacharya, S. K., Girsch, G.
2014
Cazorla, M., Brune, W. H.
2010
Stockwell, William R., Kirchner, Frank, Kuhn, Michael, Seefeld, Stephan
1997
Sokhi, Ranjeet S., Moussiopoulos, Nicolas, Baklanov, Alexander, Bartzis, John, Coll, Isabelle, Finardi, Sandro, Friedrich, Rainer, Geels, Camilla, Grönholm, Tiia, Halenka, Tomas, Ketzel, Matthias, Maragkidou, Androniki, Matthias, Volker, Moldanova, Jana, Ntziachristos, Leonidas, Schäfer, Klaus, Suppan, Peter, Tsegas, George, Carmichael, Greg, Franco, Vicente, Hanna, Steve, Jalkanen, Jukka-Pekka, Velders, Guus J. M., Kukkonen, Jaakko
2022
Thunis, P., Clappier, A., Tarrason, L., Cuvelier, C., Monteiro, A., Pisoni, E., Wesseling, J., Belis, C. A., Pirovano, G., Janssen, S., Guerreiro, C., Peduzzi, E.
2019
Fowler, David, Brimblecombe, Peter, Burrows, John, Heal, Mathew R., Grennfelt, Peringe, Stevenson, David S., Jowett, Alan, Nemitz, Eiko, Coyle, Mhairi, Liu, Xuejun, Chang, Yunhua, Fuller, Gary W., Sutton, Mark A., Klimont, Zbigniew, Unsworth, Mike H., Vieno, Massimo
2020
Manisalidis, Ioannis, Stavropoulou, Elisavet, Stavropoulos, Agathangelos, Bezirtzoglou, Eugenia
2020
Stockwell, William R., Lawson, Charlene V., Saunders, Emily, Goliff, Wendy S.
2012
Vereecken, Luc, Francisco, Joseph S.
2012
Bertram, T. H., Kimmel, J. R., Crisp, T. A., Ryder, O. S., Yatavelli, R. L. N., Thornton, J. A., Cubison, M. J., Gonin, M., Worsnop, D. R.
2011
Dallmann, Timothy R., DeMartini, Steven J., Kirchstetter, Thomas W., Herndon, Scott C., Onasch, Timothy B., Wood, Ezra C., Harley, Robert A.
2012
Dreher, David B., Harley, Robert A.
1998
Kirchstetter, Thomas W, Harley, Robert A, Kreisberg, Nathan M, Stolzenburg, Mark R, Hering, Susanne V
1999
McDonald, Brian C., Dallmann, Timothy R., Martin, Elliot W., Harley, Robert A.
2012
Harley, Robert A., Marr, Linsey C., Lehner, Jaime K., Giddings, Sarah N.
2005
Burgard, Daniel A., Bishop, Gary A., Stedman, Donald H., Gessner, Viktoria H., Daeschlein, Christian
2006
Bishop, G. A., Stedman, D. H.
1996
Bishop, Gary A., Stedman, Donald H.
2008
Ban-Weiss, George A., McLaughlin, John P., Harley, Robert A., Lunden, Melissa M., Kirchstetter, Thomas W., Kean, Andrew J., Strawa, Anthony W., Stevenson, Eric D., Kendall, Gary R.
2008
Anderson, Daniel C., Lindsay, Andrew, DeCarlo, Peter F., Wood, Ezra C.
2021
Nah, Theodora, Ji, Yi, Tanner, David J., Guo, Hongyu, Sullivan, Amy P., Ng, Nga Lee, Weber, Rodney J., Huey, L. Gregory
2018
Sanchez, Javier, Tanner, David J., Chen, Dexian, Huey, L. Gregory, Ng, Nga L.
2016
Phillips, G. J., Pouvesle, N., Thieser, J., Schuster, G., Axinte, R., Fischer, H., Williams, J., Lelieveld, J., Crowley, J. N.
2013
Palm, Brett B., Liu, Xiaoxi, Jimenez, Jose L., Thornton, Joel A.
2019
Bertram, T. H., Kimmel, J. R., Crisp, T. A., Ryder, O. S., Yatavelli, R. L. N., Thornton, J. A., Cubison, M. J., Gonin, M., Worsnop, D. R.
2011
Lee, Ben H., Lopez-Hilfiker, Felipe D., Veres, Patrick R., McDuffie, Erin E., Fibiger, Dorothy L., Sparks, Tamara L., Ebben, Carlena J., Green, Jaime R., Schroder, Jason C., Campuzano-Jost, Pedro, Iyer, Siddharth, D'Ambro, Emma L., Schobesberger, Siegfried, Brown, Steven S., Wooldridge, Paul J., Cohen, Ronald C., Fiddler, Marc N., Bililign, Solomon, Jimenez, Jose L., Kurtén, Theo, Weinheimer, Andrew J., Jaegle, Lyatt, Thornton, Joel A.
2018
Chen, Dexian, Huey, L. Gregory, Tanner, David J., Li, Jianfeng, Ng, Nga L., Wang, Yuhang
2017
Veres, P. R., Roberts, J. M., Wild, R. J., Edwards, P. M., Brown, S. S., Bates, T. S., Quinn, P. K., Johnson, J. E., Zamora, R. J., de Gouw, J.
2015
Lee, Ben H., Lopez-Hilfiker, Felipe D., Mohr, Claudia, Kurtén, Theo, Worsnop, Douglas R., Thornton, Joel A.
2014
Albrecht, Sascha R., Novelli, Anna, Hofzumahaus, Andreas, Kang, Sungah, Baker, Yare, Mentel, Thomas, Wahner, Andreas, Fuchs, Hendrik
2019
Lindsay, Andrew J., Wood, Ezra C.
2022
Bowers, Bailey B., Thornton, Joel A., Sullivan, Ryan C.
2023
Riedel, Theran P., Lang, Johnsie R., Strynar, Mark J., Lindstrom, Andrew B., Offenberg, John H.
2019
Robinson, Michael A., Neuman, J. Andrew, Huey, L. Gregory, Roberts, James M., Brown, Steven S., Veres, Patrick R.
2022
Unknown
Sulbaek Andersen, M. P., Nielsen, O. J., Hurley, M. D., Ball, J. C., Wallington, T. J., Ellis, D. A., Martin, J. W., Mabury, S. A.
2005
Bowers, Bailey B., Thornton, Joel A., Sullivan, Ryan C.
2023
Riedel, Theran P., Lang, Johnsie R., Strynar, Mark J., Lindstrom, Andrew B., Offenberg, John H.
2019
Vyas, Sandhya M., Kania-Korwel, Izabela, Lehmler, Hans-Joachim
2007
US EPA, OW
2021
Unknown
Unknown
Riedel, Theran P., Lang, Johnsie R., Strynar, Mark J., Lindstrom, Andrew B., Offenberg, John H.
2019
Faust, Jennifer A.
2023
Jahnke, Annika, Ahrens, Lutz, Ebinghaus, Ralf, Berger, Urs, Barber, Jonathan L., Temme, Christian
2007
Ahrens, Lutz, Shoeib, Mahiba, Harner, Tom, Lane, Douglas A., Guo, Rui, Reiner, Eric J.
2011
Jahnke, Annika, Ahrens, Lutz, Ebinghaus, Ralf, Temme, Christian
2007
Johansson, J. H., Salter, M. E., Navarro, J. C. Acosta, Leck, C., Nilsson, E. D., Cousins, I. T.
2019
Admin, ITRC
Karásková, Pavlína, Codling, Garry, Melymuk, Lisa, Klánová, Jana
2018
Paragot, Nils, Bečanová, Jitka, Karásková, Pavlína, Prokeš, Roman, Klánová, Jana, Lammel, Gerhard, Degrendele, Céline
2020
Yu, Nanyang, Wen, Haozhe, Wang, Xuebing, Yamazaki, Eriko, Taniyasu, Sachi, Yamashita, Nobuyoshi, Yu, Hongxia, Wei, Si
2020
Yamazaki, Eriko, Taniyasu, Sachi, Wang, Xinhong, Yamashita, Nobuyoshi
2021
Barber, Jonathan L., Berger, Urs, Chaemfa, Chakra, Huber, Sandra, Jahnke, Annika, Temme, Christian, Jones, Kevin C.
2007
Jahnke, Annika, Barber, Jonathan L., Jones, Kevin C., Temme, Christian
2009
Arp, Hans Peter H., Goss, Kai-Uwe
2008
Zhang, Mengke, Yamada, Kyle, Bourguet, Stephen, Guelfo, Jennifer, Suuberg, Eric M.
2020
Cheng, Peiyang, Pour-Biazar, Arastoo, Wu, Yuling, Kuang, Shi, McNider, Richard T., Koshak, William J.
2024
Liu, Jiachen, Chen, Eric, Capps, Shannon L.
2024
Liu, Yanhui, Li, Jiayin, Ma, Yufang, Zhou, Ming, Tan, Zhaofeng, Zeng, Limin, Lu, Keding, Zhang, Yuanhang
2023
He, Xiao, Zheng, Xuan, Guo, Shuwen, Zeng, Lewei, Chen, Ting, Yang, Bohan, Xiao, Shupei, Wang, Qiongqiong, Li, Zhiyuan, You, Yan, Zhang, Shaojun, Wu, Ye
2024
De Haan, David O., Hawkins, Lelia N., Weber, Jacob A., Moul, Benjamin T., Hui, Samson, Cox, Santeh A., Esse, Jennifer U., Skochdopole, Nathan R., Lynch, Carys P., De Haan, Audrey C., Le, Chen, Cazaunau, Mathieu, Bergé, Antonin, Pangui, Edouard, Heuser, Johannes, Doussin, Jean-François, Picquet-Varrault, Bénédicte
2024
Unknown
Höpfner, Michael, Volkamer, Rainer, Grabowski, Udo, Grutter, Michel, Orphal, Johannes, Stiller, Gabriele, von Clarmann, Thomas, Wetzel, Gerald
2016
Wang, Yuwei, Li, Chuang, Zhang, Ying, Li, Yueyang, Yang, Gan, Yang, Xueyan, Wu, Yizhen, Yao, Lei, Zhang, Hefeng, Wang, Lin
2024
Gaston, Cassandra J., Prospero, Joseph M., Foley, Kristen, Pye, Havala O. T., Custals, Lillian, Blades, Edmund, Sealy, Peter, Christie, James A.
2024
Plauchu, Robin, Fortems-Cheiney, Audrey, Broquet, Grégoire, Pison, Isabelle, Berchet, Antoine, Potier, Elise, Dufour, Gaëlle, Coman, Adriana, Savas, Dilek, Siour, Guillaume, Eskes, Henk
2024
Krol, Maarten, van Stratum, Bart, Anglou, Isidora, Boersma, Klaas Folkert
2024
Nürnberg, Pinchas, Rettinger, Markus, Sussmann, Ralf
2024
A Blog:
Lee's Climate Pointers for non-Chemists(from a Chemistry Perspective)

All the Rage
and Global Warming Potentials
This story begins with the coldness of space .
Because it's really hard to live in the cold. Imagine Paris, Vienna, and Edinburgh had similar climates to Antarctica. We would be missing out on some pretty good philosophy, right? Culture as we know it would be pretty bland, if existent at all.
Our universe kind-of prefers coldness, so galaxies & their planets need to be pretty nifty in their attempt to stay warm. We generally say cold things have less energy than their hot counterparts. Energy is expressed in movement and light (among other things), and high movement or brightness among matter is correlated to high temperature.
If we put a big rock in space and shine light (energy) on it, the rock will get rid of the energy! The inbound energy flux (energy of light on the exposed surface) is always equal to the outbound energy flux. This phenomenon is called radiative balance 1 and it's a really troubling physical principle for anyone interested in living on big rocks in space. Right? Space takes all objects' outbound energy. When something is taking energy from something else, it is cold.
Boring, random rocks/objects are not stars, correct. But they do give off energy in the form of light! They abide by the principle of radiative balance.
It's important to remember not all light is visible to us. Earth gives off mainly infrared light (IR is invisible to us). 2
You know how walking down an empty stairwell compels you, by the overpowering force of immaturity, to pull from the depths of your throat a big "hoooot"? And how if you pull out the "hoooot" note just right, a proper shake buzzes about the air? We call this resonance. 3
Earth's outward infrared light happens to have a particularly interesting property: it tickles some molecules' fancies. To be technical, this light causes certain molecules to resonate! This incredible video shows just this.
When light resonates with the vibration of molecules, the molecules move faster... meaning their temperature increases ! (Temperature is the average energy of movement.)
Carbon dioxide, water, methane are a few of the molecules that resonate with infrared radiation and absorb it. Without these compounds, Earth would give away energy without warming up. (Earth would be a cold, cold place lacking any good philosophy!)
This is important. The resonating compounds are very good at what they do. More of them $\rightarrow$ more absorbed infrared light $\rightarrow$ higher temperatures.
These compounds are what maintain a temperature of roughly $15\ ^oC/60\ ^oF$ close to Earth's surface. What they don't absorb gets emitted! (Because of radiative balance.)
Goldilocks needed a bowl of porridge with a temperature that was just right. Life as we know it (and good philosophy) also needs Earth stay at a temperature that is just right.
Carbon dioxide is:
Each dashed line in the image below corresponds to an object emitting infrared light. These objects are called black bodies and the profile of the light they emit is characteristic of their temperature. 4 The area under their curve is unique. Black bodies are ideal objects. This is why their curves look so perfect.
The solid black line represents Earth trying to be a black body. Clearly, it's not doing a great job. The plot shows measurements of infrared light that Earth is emitting from the Sahara Desert. We can see that there are certain "wavelengths" (top axis) of light that we don't see much of. In other words, there are some big dips. (Like at $15 \ \mu m$, $10.5 \ \mu m$, and $8-7.5 \ \mu m$.)
Those dips represent infrared light emitted by Earth that did not reach the measurement device. What happened to it? The resonating molecules (colloquially, greenhouse gasses ) absorbed the light before got there!
Because of radiative balance ! All of that infrared radiation is being absorbed by (and thus heating) the resonating molecules. Because of this, our atmosphere heats up. More resonance $\rightarrow$ higher baseline of Earth's curve in this plot $\rightarrow$ warmer atmosphere.
This visualization hopefully makes clear:
1
Radiative balance of Earth. "The only source of heat on Earth is solar radiation. As any physical body with a temperature above absolute zero, Earth loses heat in the form of infrared radiation proportional to the fourth power of its absolute temperature. The Earth as a planet is in almost perfect thermal steady state and therefore the top of the atmosphere must be in a complete globally-averaged radiative balance." -
Heat Transport, Oceanic and Atmospheric
2
IR radiation. "That portion of the
electromagnetic spectrum
that extends from the long wavelength, or red, end of the visible-light range to the microwave range." -
Britannica
3
Resonance in chemicals. "When the frequency of an external oscillation or vibration matches an object (or cavity’s)
natural frequency
, and as a result either causes it to vibrate or increases its amplitude of oscillation." -
SCIENCING
4
Black body curves. "At thermodynamic equilibrium, the rate at which an object absorbs radiation is the same as the rate at which it emits it. Therefore, a good absorber of radiation (any object that absorbs radiation) is also a good emitter. A perfect absorber absorbs all electromagnetic radiation incident on it; such an object is called a black body." -
Physics LibreTexts
It's important to remember the global atmosphere is a very dynamic system. Oftentimes, in such systems, our human tendency to make predictions based off common intuition leads us astray. The sky doesn't always work as we think it will. In this blog, I will present a few examples of atmospheric alterations that have competing effects. My goal is to show that:
Let's categorize two ways an atmospheric component can impact the environment:
Sulfur dioxide ($SO_2$) is an air pollutant. It's a dangerous gas to human, animal, and plant health and contributes to acid rain because it is the source of sulfuric acid ($H_2SO_4$) in the atmosphere. On the other hand, the presence of $H_2SO_4$ accelerates particle formation and growth in the sky. These particles reflect sunlight back to space, which effectively cools the planet. ( See my blog post about light and Earth's temperature ).
So, does adding $SO_2$ to the sky have a net negative or net positive on the planet? This seems like a difficult metric to quantify.
1
EPA defines GWP to be equivalent magnitude of warming a unit amount of substance will have over 100 years as compared to the same amount in $CO_2$ -
Atmospheric Lifetime and Global Warming Potential Defined

Lee Feinman is a self-proclaimed expert on nothing. This is why he studies chemistry (to become an expert at chemistry) and writes essays, poems, and short stories about the journeys taken by he who is in search of expertise. He is a native of Media, PA. Aside from pursuing a professional career in atmospheric chemistry, Lee enjoys a literary and philosophical interest in reading & writing in his free time.
My Selected Essays
Books I Have Read and Like to Talk About
The Projects I Have Worked On
I've got a lot to say here! I love opening back up books I've read and remembering exactly why I loved them. Contemplating why some amazing human thought such thoughts and how they organized that thought in such a way.
I just haven't gotten there yet.
In the meantime... feel free to keep tabs on my cycling (below)! The gaps in time are due to forgetting to press "start" on Strava. Definitely .ot because I'm lazy and don't ride a lot...
Here's a list of the things I've done (definitely not exhaustive!):
# in the terminal:
cd DirectoryToWebsiteFiles
python script.py
